Rainbow proviral HIV-1 DNA digital PCR assay
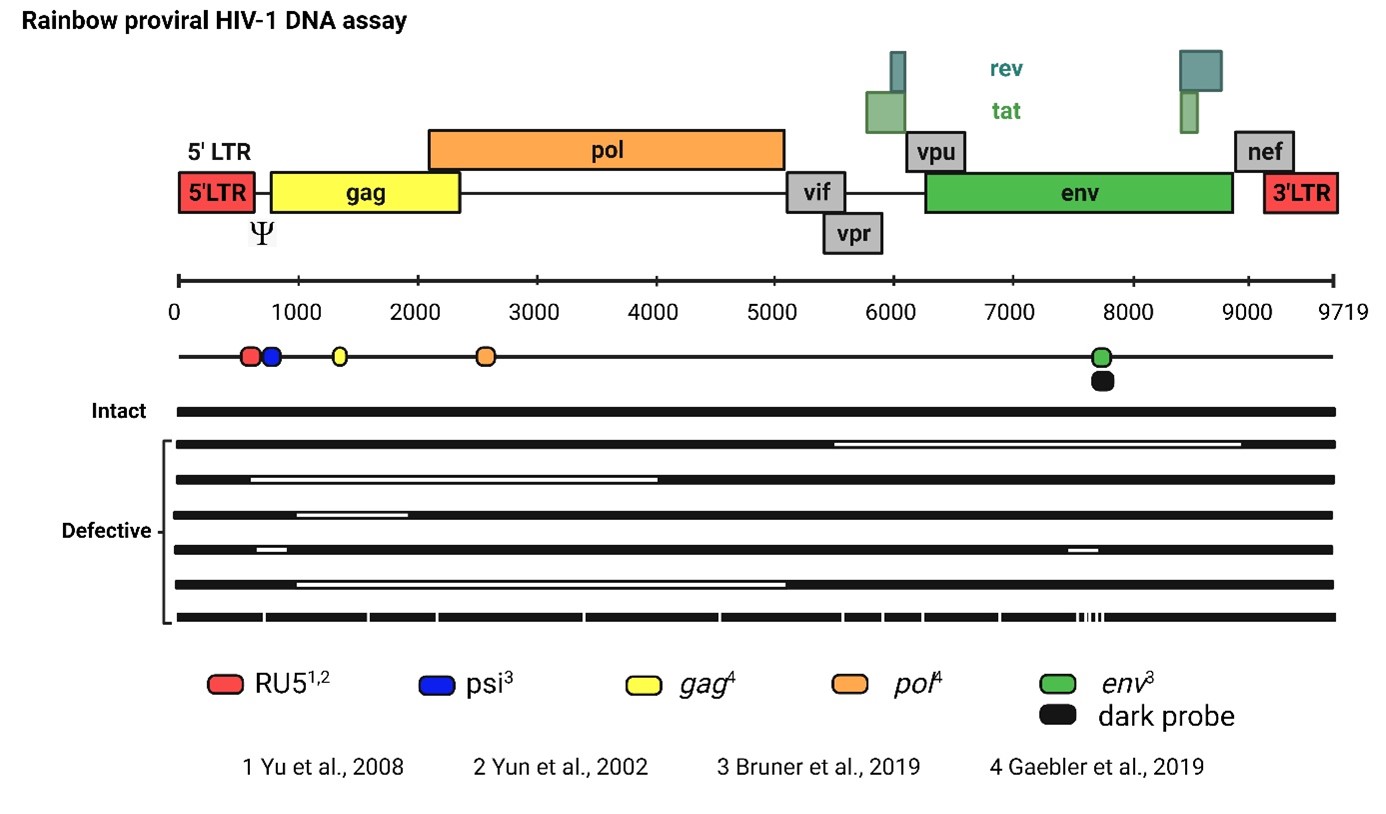
Figure 1: Schematic overview of the Rainbow proviral HIV-1 DNA dPCR assay.
The five target regions used in this assay are displayed at their respective locations in the HIV-1 genome, with a different color for each region. RU5 is used to quantify total HIV-1 DNA, whereas the other four regions (psi, gag, pol and env) are used to quantify intact HIV-1 DNA. A dark probe without fluorescent dye is used at the same location of env to bind hypermutated env (competition probe).
Persistent latent reservoirs of intact HIV-1 proviruses, capable of rebounding despite suppressive ART, hinder efforts towards an HIV-1 cure. Hence, assays specifically quantifying intact proviruses are crucial to assess the impact of curative interventions. Two recent assays have been utilized in clinical trials: intact proviral DNA assay (IPDA) and Q4PCR. While IPDA is more sensitive due to amplifying short fragments, it may overestimate intact fractions by relying only on quantification of two proviral regions. Q4PCR samples four proviral regions, yet is sequencing-based, favoring amplification of shorter, hence non-intact, proviral sequences. Leveraging digital PCR (dPCR) advancements, we developed the ‘Rainbow’ 5-plex proviral HIV-1 DNA assay, assessing it with standard materials and samples from 83 people living with HIV-1. The Rainbow assay proved equally sensitive but more specific than IPDA, is not subjected to bias against full-length proviruses, enabling high-throughput quantification of total and intact reservoir size. This innovation offers potential for targeted evaluation and monitoring of potential rebound-competent reservoirs, contributing to HIV-1 management and cure strategies.
More information:
biorxiv.org/content/10.1101/2023.08.18.553846v1
Contact:
mareva.delporte@ugent.be
linos.vandekerckhove@ugent.be

WERELDWIJD LEVEN 37 MILJOEN MENSEN MET HIV. STEUN HET FONDS HIV ONTRAFELEN en Help hen.
-
Of via overschrijving:
Universiteitsfonds UGent - IBAN BE26 3900 9658 0329
- Met duidelijke vermelding: "Fonds HIV"
- Giften zijn voor 45% fiscaal aftrekbaar vanaf 40 euro op jaarbasis.
- Meer info over hoe je het HIV onderzoek kan steunen.